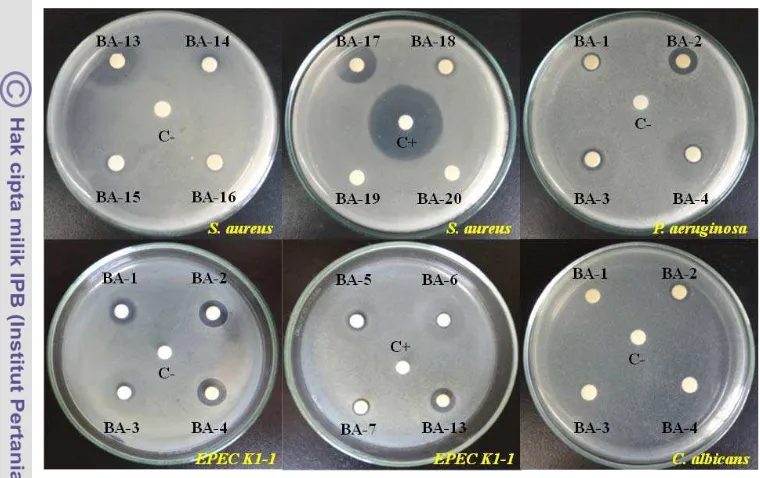
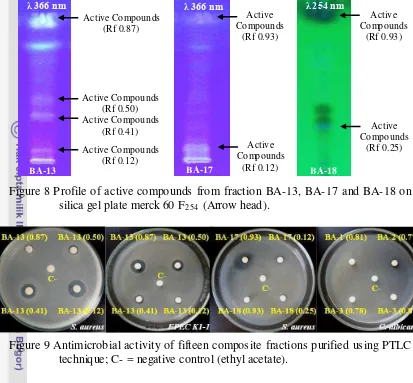
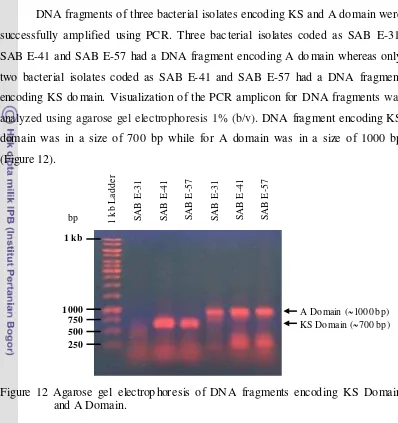
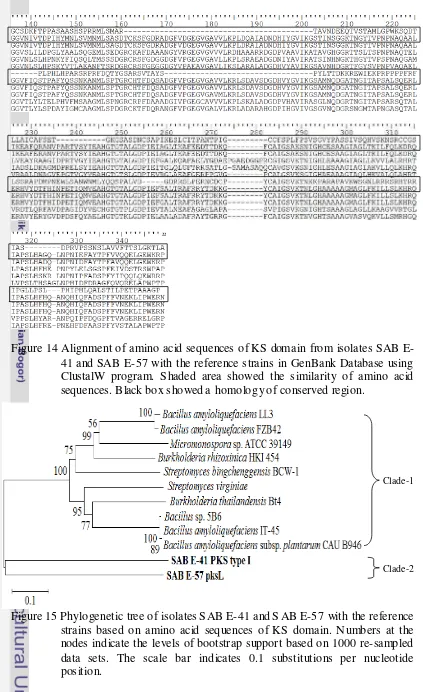
Antimicrobial Activity of Bioactive Compounds Isolated from Marine Bacteria Associated with Sponge Jaspis sp. and Their Genetics Analys is.
Teks penuh
Gambar


Dokumen terkait
Pembalikkan telur yang biasanya dilakukan oleh peternak satu persatu setiap pagi, siang, dan malam kemudian juga ada yang dengan menggerakan bak telur beberapa derajat
Tujuan penelitian ini untuk mengetahui apakah terdapat hubungan antara higiene perseorangan, sanitasi lingkungan dan status gizi dengan kejadian skabies pada
Hasil ini lebih tinggi dari penelitian Mukai et al yang melaporkan bahwa kadar gula darah puasa terganggu atau Impaired Fasting Glucose (IFG) berisiko 3,49 kali untuk terjadinya
Simpulan dari penelitian yaitu bahwa pemberian air rebusan kunyit sebagai suplemen kesehatan pada air minum ayam broiler sampai taraf 75% mampu mempertahankan profil sel
Pada permasalahan optimasi sumber daya, baik itu minimalisasi biaya ataupun maksimalisasi keuntungan, yang bersifat linear, dapat dibuat sebuah tool yang berguna untuk
Dari data peringkat penyakit kanker pasien rawat inap di Rumah sakit di Indonesia tahun 2005, Carcinoma nasopharynx berada pada peringkat 10 (3,4 %)1. Penelitian ini
Penataan ruang Kawasan Taman Nasional Lorentz bertujuan untuk mewujudkan pelestarian kawasan taman nasional lorentz sebagai salah satu pusat konservasi keanekaragaman hayati dan
Budidaya dengan teknologi pencultur dapat memanfaatkan lahan budidaya di perairan pesisir yang dangkal hingga ke dalam perairan di perairan dalam dengan biota budidaya